The ABC-Myxo browsing Table provides enhanced functionality for streamlined browsing. Users can view information related to Biosynthetic Gene Clusters (BGCs) or Gene Cluster Families (GCFs) by navigating between the tabs at the top of the page. Below are descriptions of the key columns for each tab.
All Biosynthetic Gene Clusters (BGCs) Tab
- BGC Type: Indicates the predicted type of BGC as determined by antiSMASH.
- Completeness: Specifies whether the BGC is complete or fragmented.
- MIBiG: Identifies whether this BGC matches a record in the MIBiG database.
- GCF ID: Displays the Gene Cluster Family (GCF) to which this BGC belongs. Clicking on the GCF link redirects to the corresponding GCF page.
- Source: Denotes the source genome from which the BGC is derived.
- Strain: Provides a link to the NCBI database for downloading the genome.
Clicking on the name of any BGC will redirect to the associated antiSMASH results page.
Grouped by Gene Cluster Family (GCF)
- BGC Count: Displays the number of BGCs within the GCF.
- MIBiG: Lists the matched MIBiG BGCs within the GCF.
- Source: Indicates the sources of the genomes from which the BGCs in this GCF originate.
- GCF Type: Describes the type of BGCs grouped within this GCF.
- Cytoscape: Provides a link to the Cytoscape clustering visualization page for this GCF.
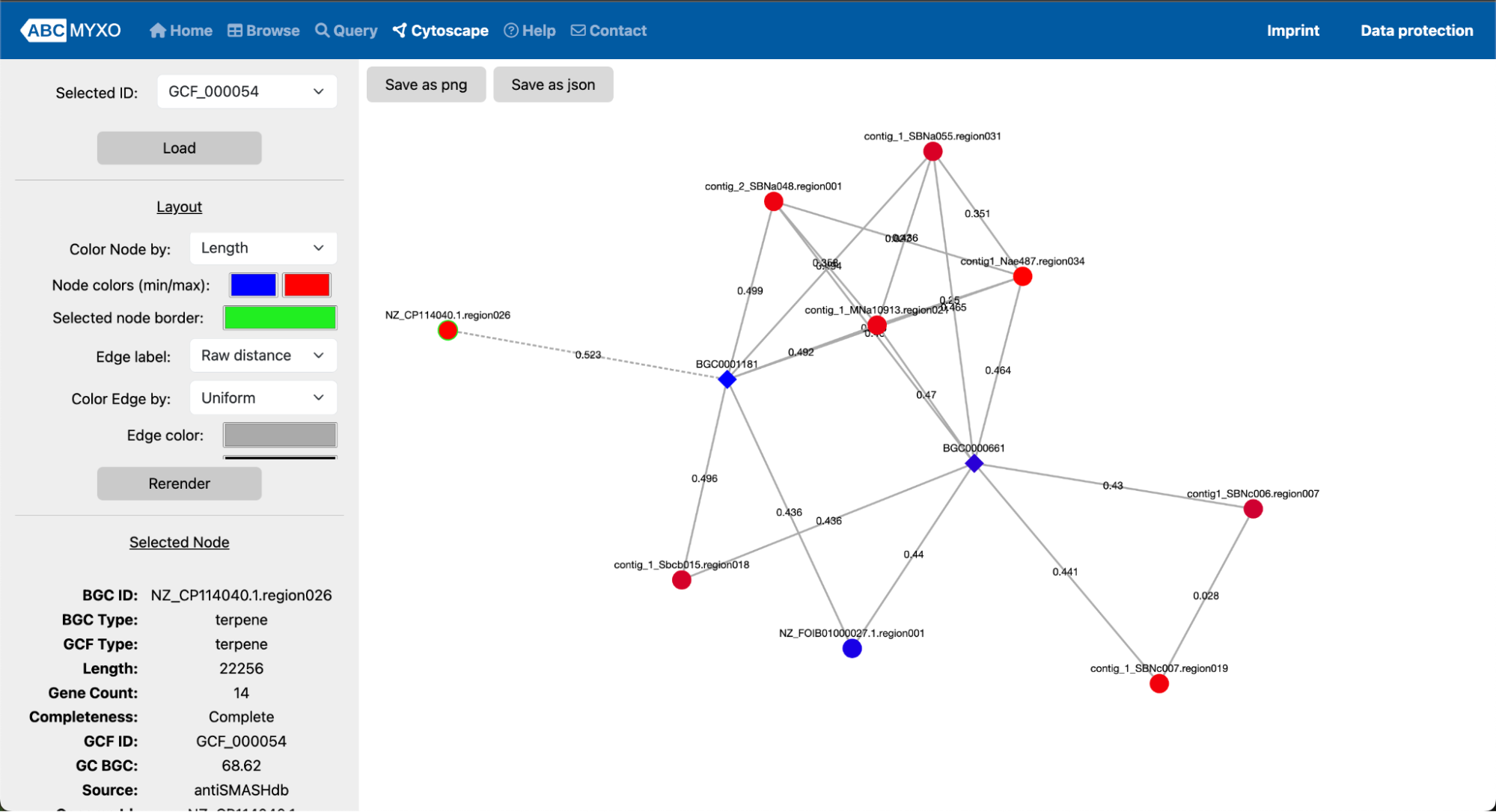
While on the Cytoscape clustering visualization page (Users can also directly click the ‘Cytoscape’ section at the top of the header), users can customize the layout by adjusting parameters within the “Layout” panel located on the left side of the page. Each parameter can be modified using a drop-down menu. After making changes, click the “Load” button at the top of the page to refresh the visualization. Additionally, nodes can be dragged to a new position, and the layout can be re-rendered by clicking the “Rerender” button at the bottom of the page. Clicking on a specific node, which represents a BGC, will display detailed information in the “Selected Node” area located in the bottom-left corner of the page.
ABC-Myxo is equipped to search for homologous genes and BGCs of the ABC-Myxo database with users’ input sequences. Users can find these two functionalities by clicking the ‘Query’ menu at the top.
BLAST: Search for homologous genes
Users can submit their own sequences and choose from different modules (“BLASTn”, “BLASTp” or “tBLASTn”) to perform the homologous genes search. Additionally, users can adjust the “Maximum number of results” and “E-value cutoff” parameters. Once the job starts, you are given a job ID where you can use it as a reference to retrieve the results in the “Results of the Existing Job” section. An example of the job (BLASTn) can be seen as follows:
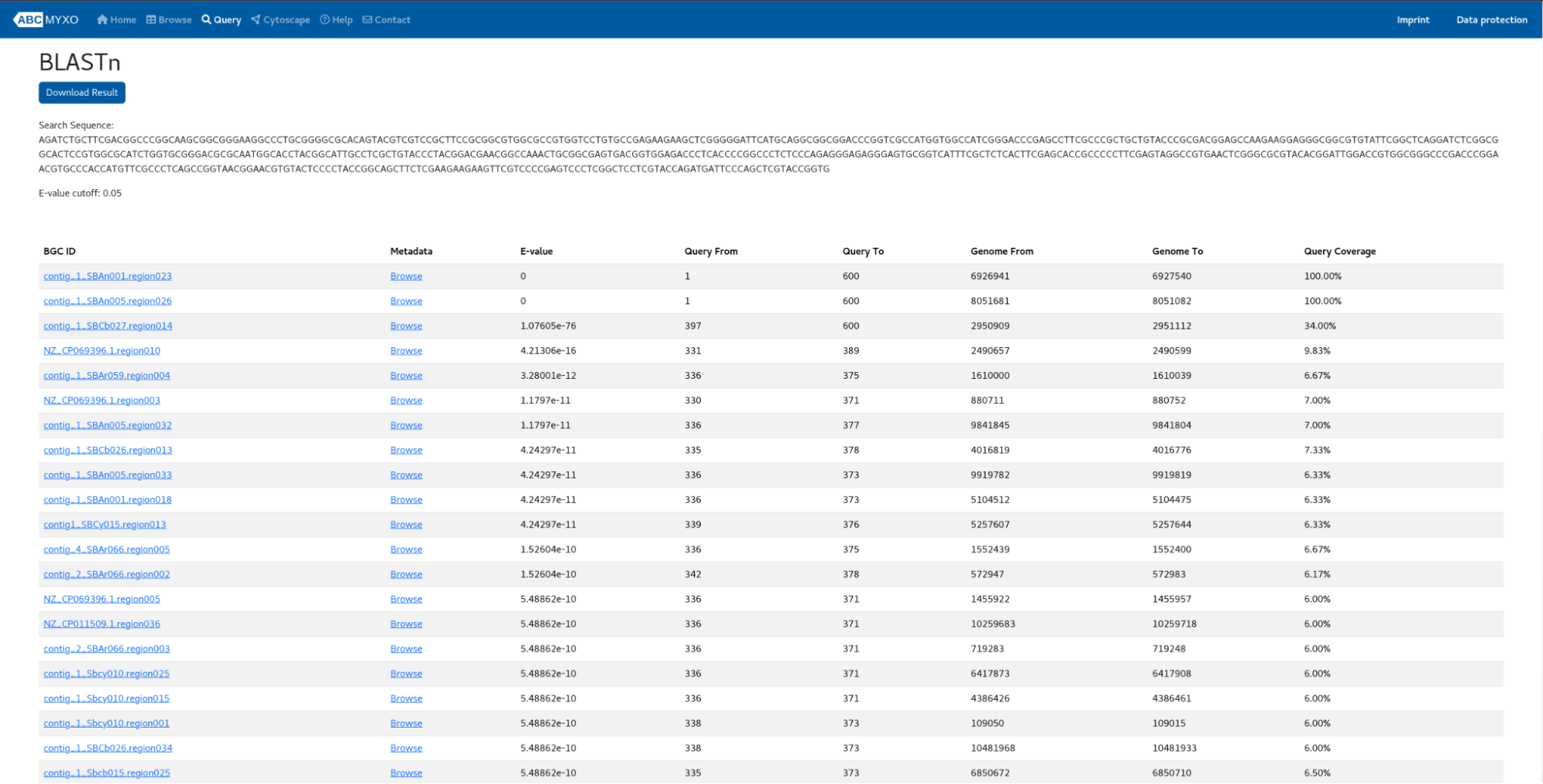
For each hit, the corresponding BGC ID, the metadata (GCF), E-value, matching query starting and ending coordinates, matching subject genome starting and ending coordinates, and query coverage are given. Users can download the results as a CSV file at the top of the page.
cblaster: Search for homologous BGCs
Users can also submit the whole BGC as a query to search for the homologous BGCs from the ABC-Myxo database. To do so, users can upload their own BGCs in gbk format using ‘Choose File’ button. After submitting the job, similarly, an ID is given to each job, which can be used to view the results at any time as explained above in the blast section. An example of the cblaster results can be shown as follows:
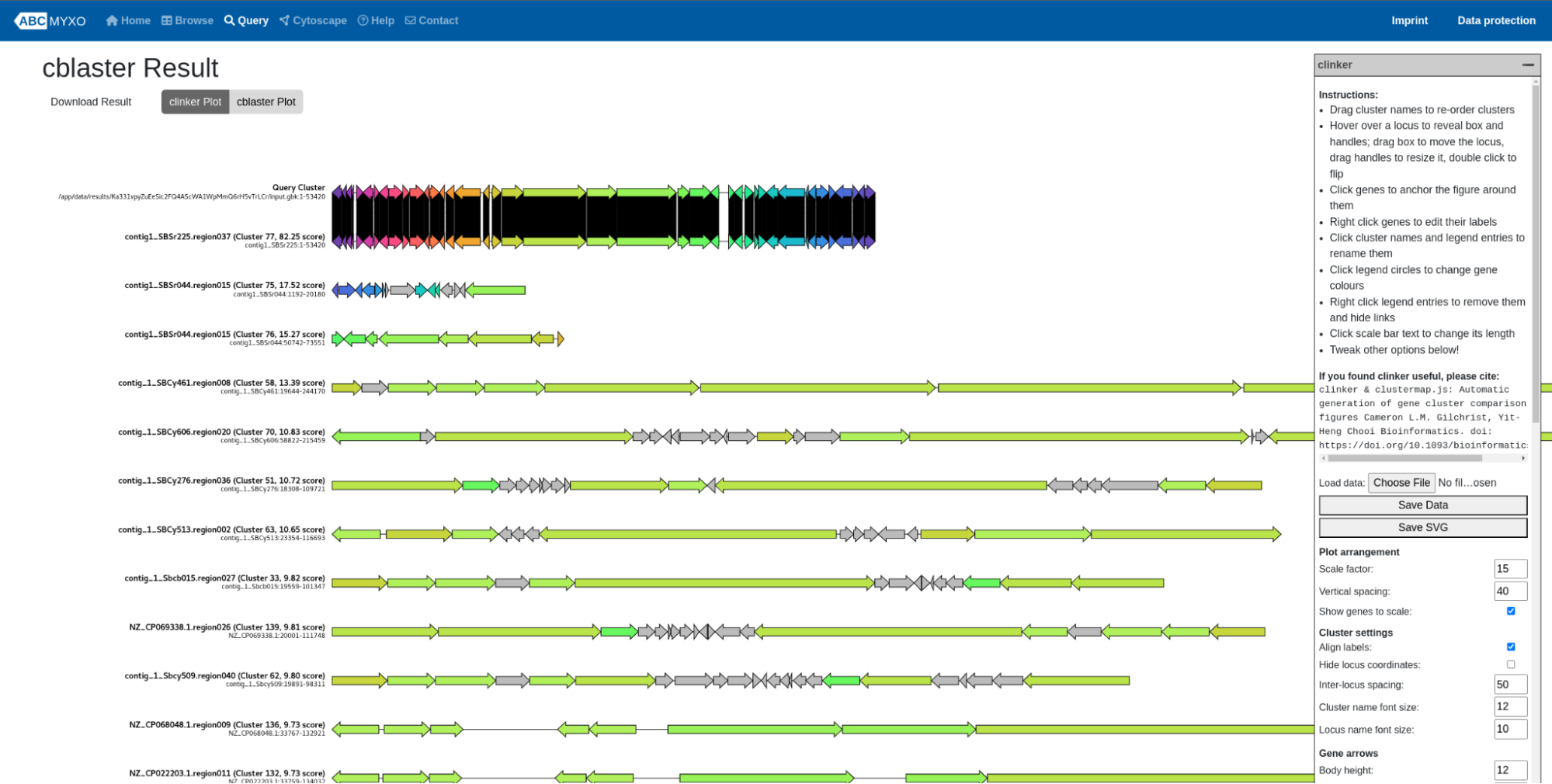
Cblaster outputs the similar BGC hits of the query BGCs. Users can choose using the visualization between ‘clinker Plot’ and ‘cblaster Plot’. The instructions of each plot can be found on the right side of the corresponding page.