How to use StructMAn Web
Select a topic you want to learn
-
How to upload and submit your sequences
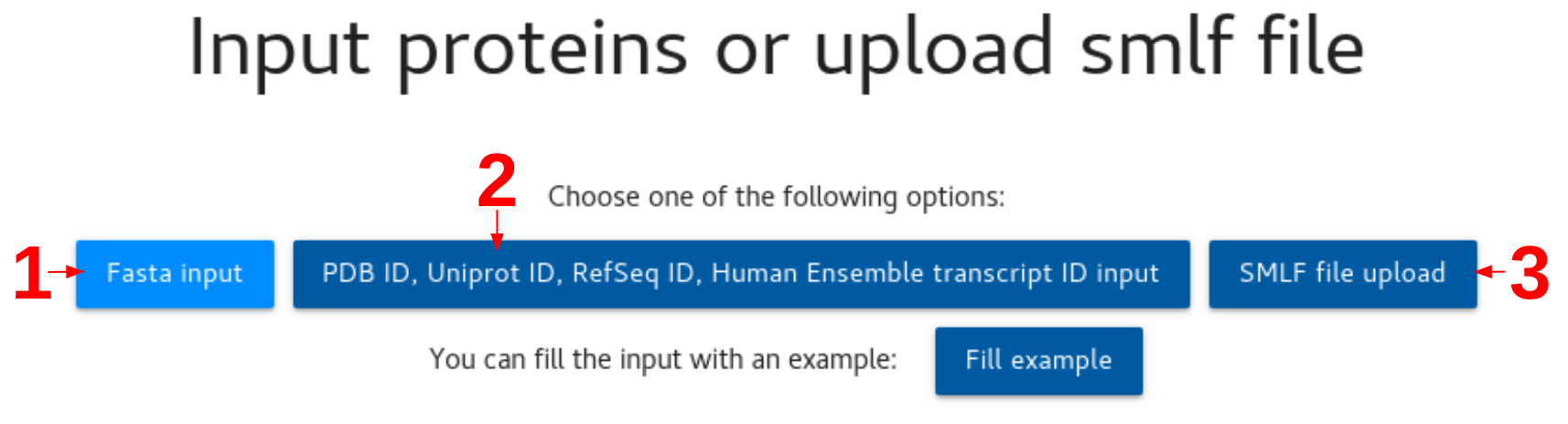
You can input your sequences using one of the following methods:
1. Amino acid sequence (FASTA format)
2. PDB ID, Uniprot ID, RefSeq ID, Human Ensemble transcript ID
3. Upload SMLF (simple mutation list format) file, you can find information here: SMLF - Structman docs
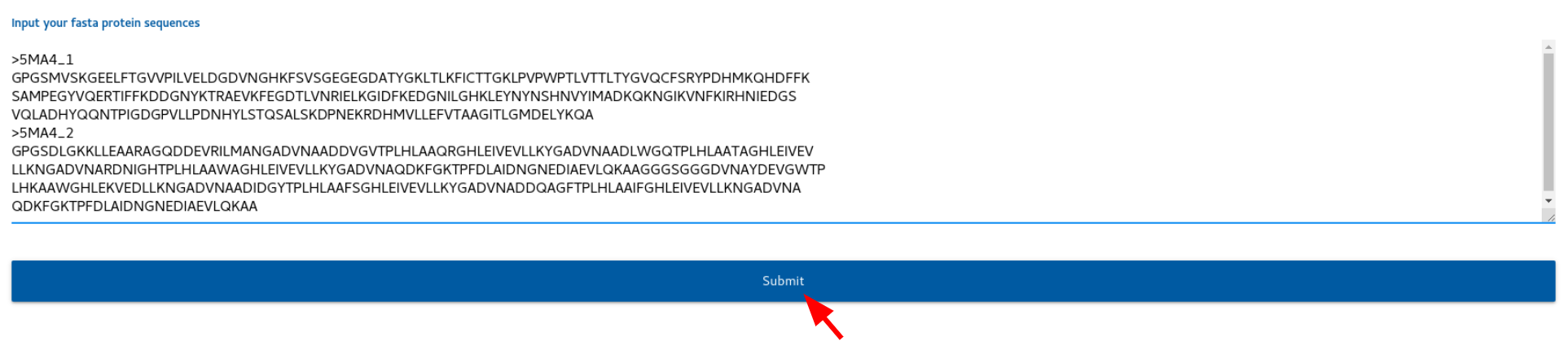
After you enter your sequences, click submit button
-
How to check your results
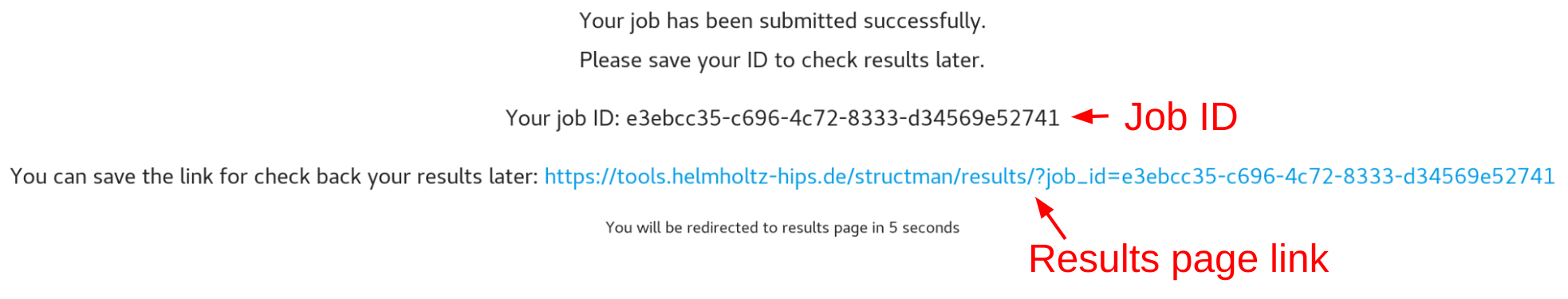
After you uploaded your sequences, you will be given your job id and results page link
You will be automatically redirected to your results page
Or you can save your Job ID to check your results later
You can check & download your result in results page
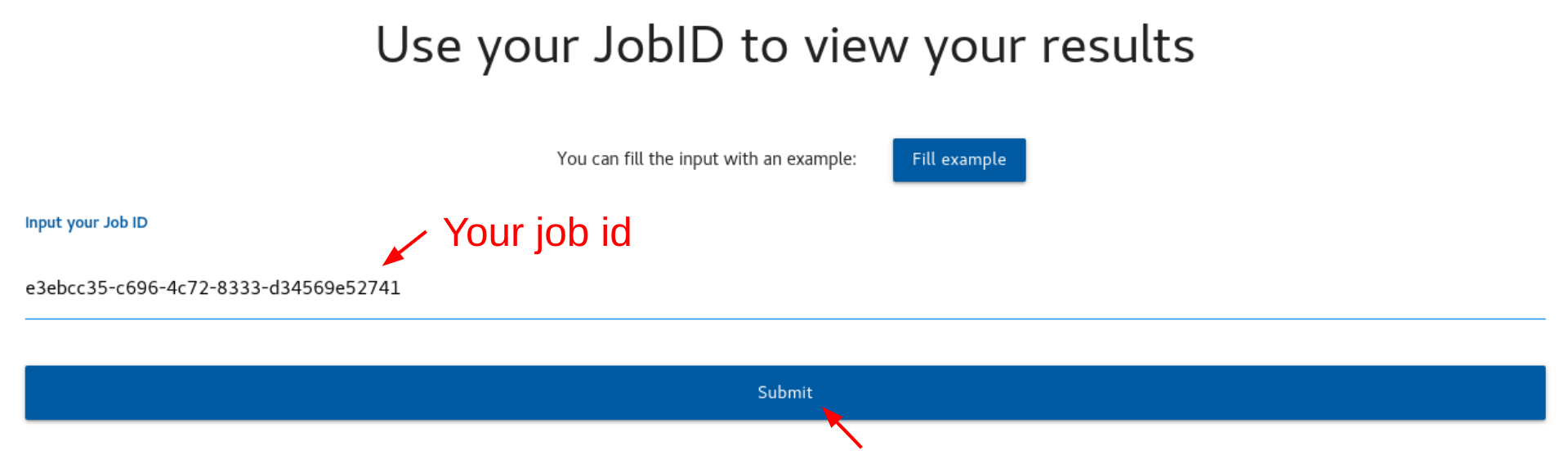
To view and download your results, please follow these steps:
1. Enter your job ID in the provided input field.
2. Click the "Submit" button.
3. Wait for the results to load. This may take a few moments depending on the size of your data.
-
Interpret your results
Results are served in 3 different containers.

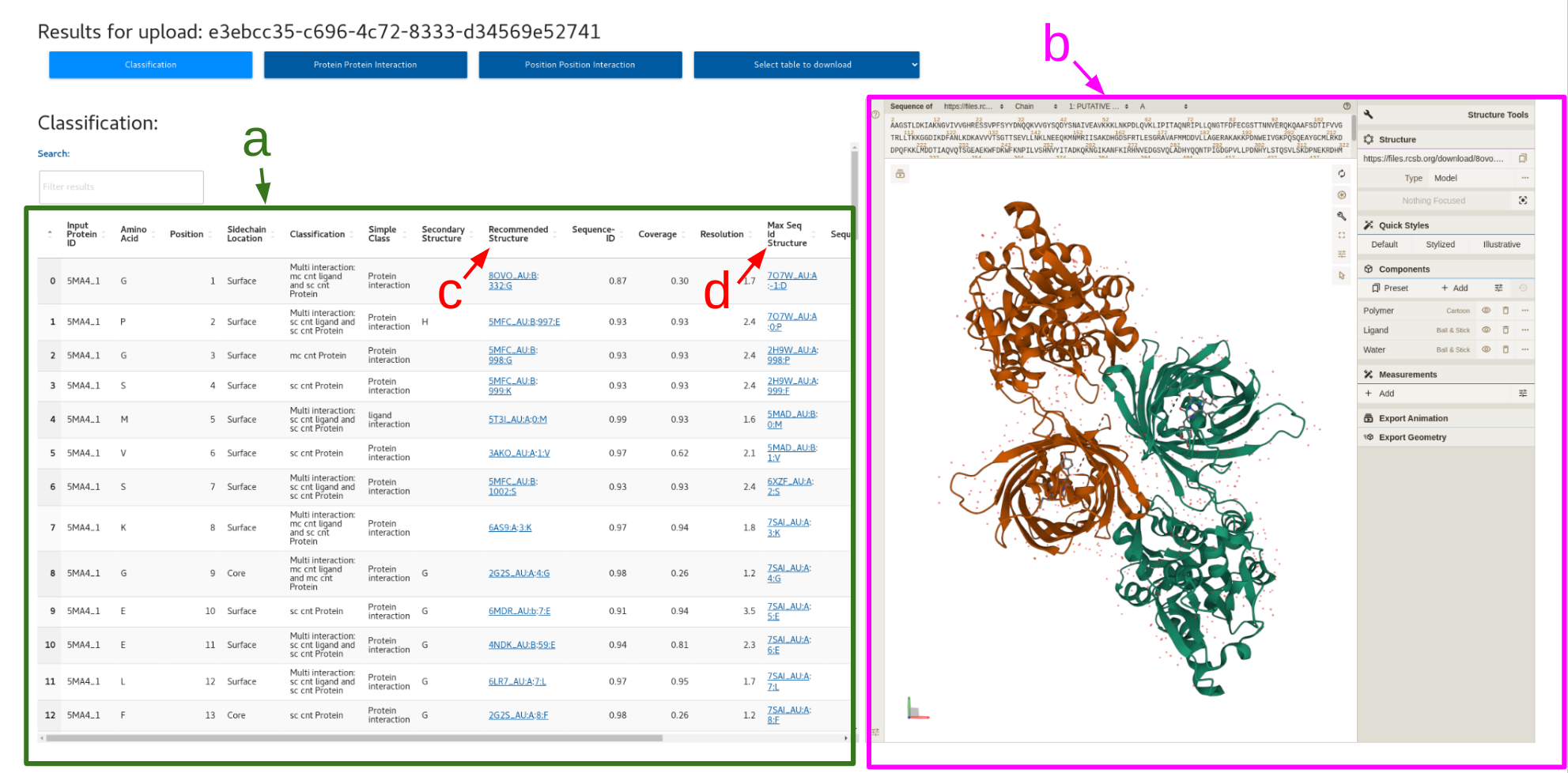
1. Classification
a. Classification table
b. PDB viewer
c. Clickable recommended structure column
d. Clickable max seq id structure column
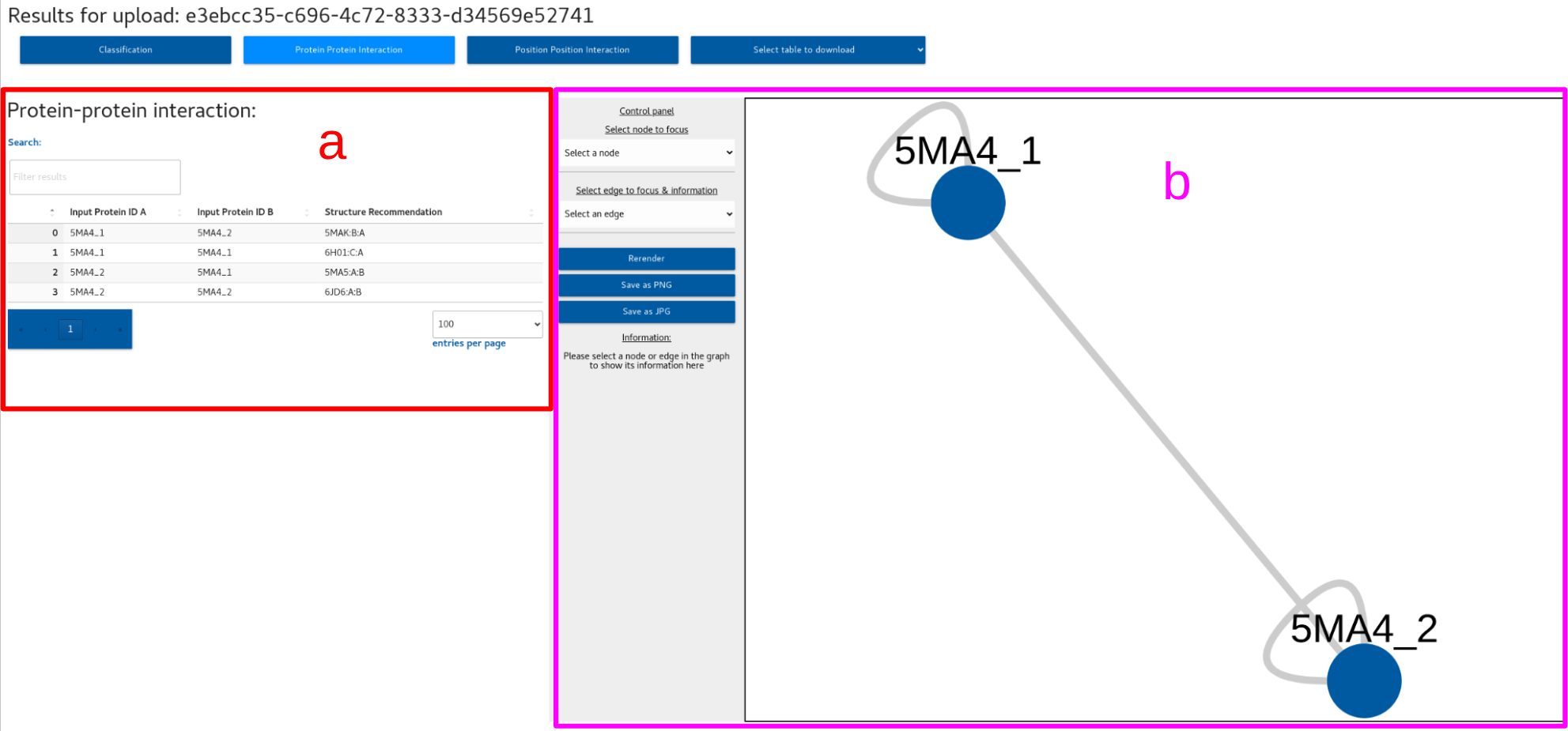
2. Protein protein interaction
a. Protein protein interaction table
b. Network viewer
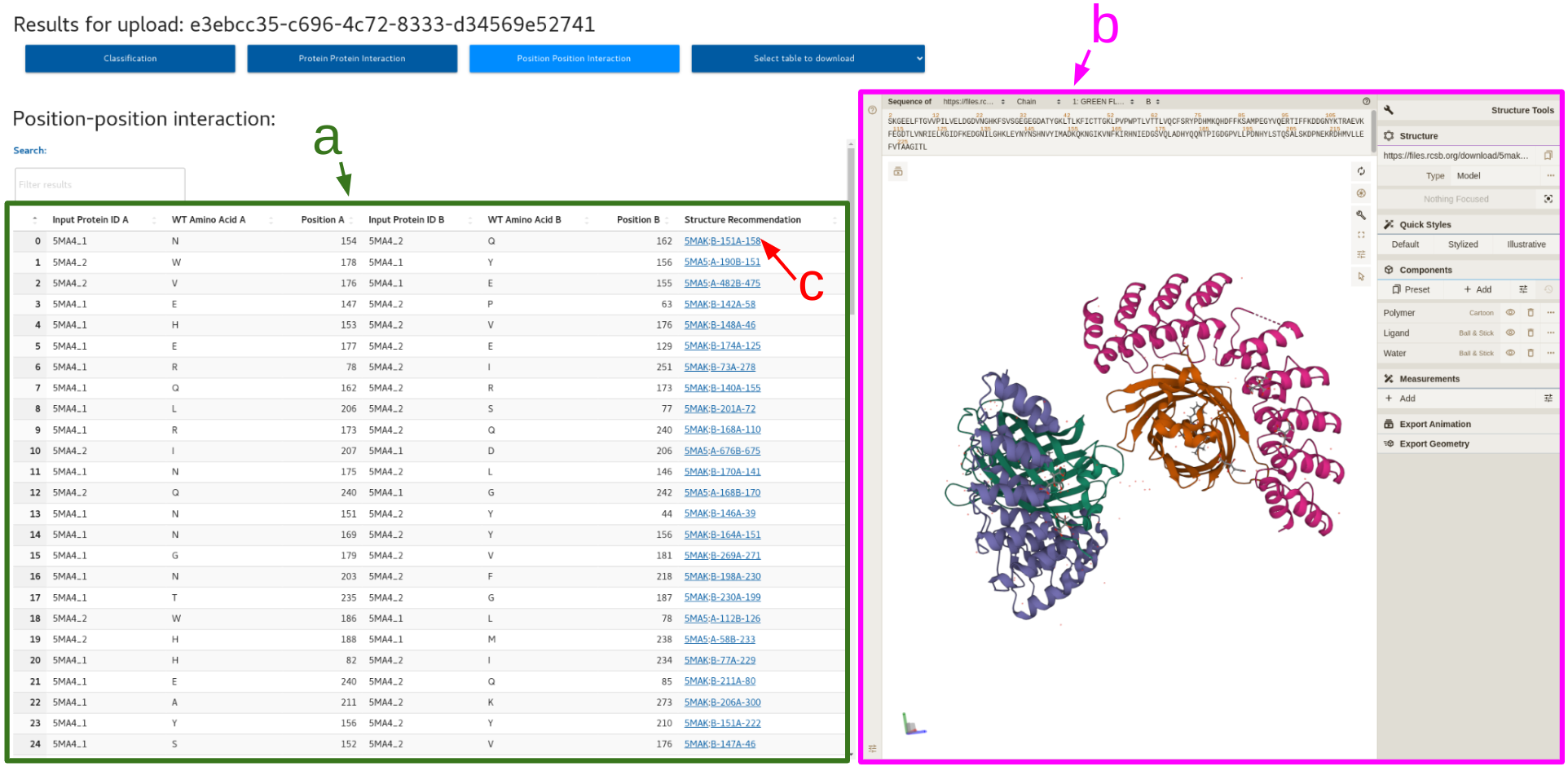
3. Position position interaction
a. Position position interaction table
b. PDB viewer
c. Clickable structure recommendation column
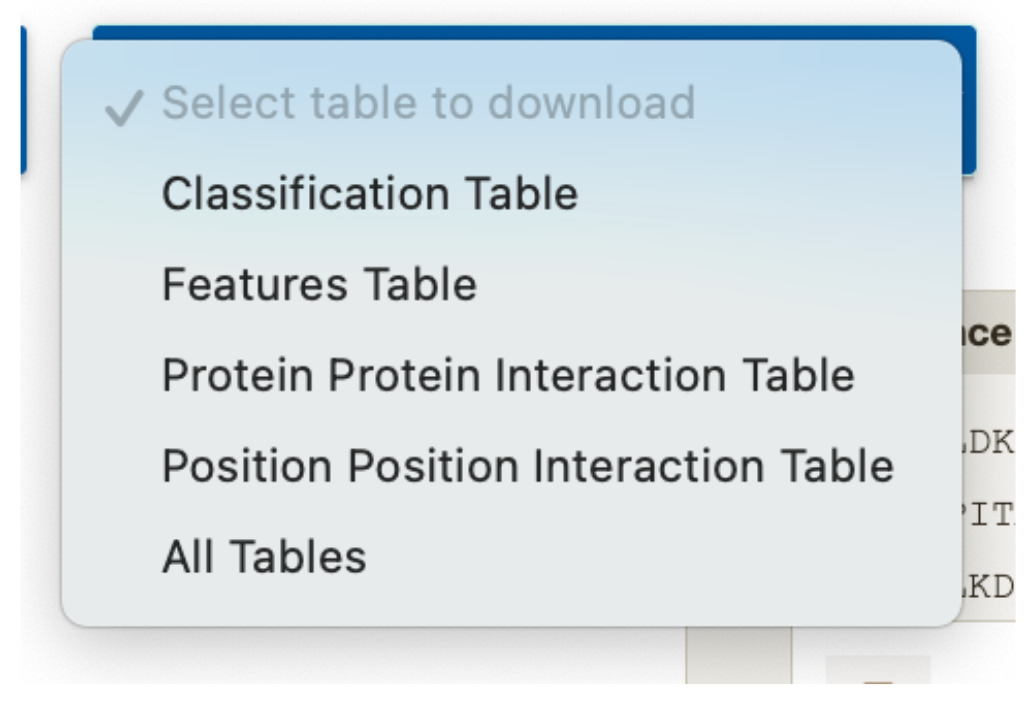
To download tables, you can use the select table to download button.
You can select individual or all the tables to download